Healthy Microbiome is Essential to Optimize Health
TNO develops better research tools that enable the development of Microbiome-Directed Health Products.
Application areas are,
Drug Metabolism by Intestinal Bacteria
The metabolism of pharmaceuticals by intestinal bacteria is one of the crucial factors influencing the effectiveness and side effects of drugs. Intestinal bacteria can metabolize drugs into different compounds, potentially impacting the pharmacokinetics and mechanisms of action of the drugs. The following are some key points related to this issue:
● Prodrug Activation: Some drugs are designed as "prodrugs" that are activated by specific bacteria in the intestine. These drugs need to be converted by certain bacteria in the intestine before being transformed into effective drugs in the body.
● Diversity of Intestinal Bacteria: The composition of individuals' intestinal microbiota varies, with differences in the types and quantities of different bacteria. This diversity may lead to varying drug metabolism profiles among individual patients, even when taking the same drug.
● Generation of By-Products: During the process of drug metabolism by intestinal bacteria, various metabolites may be produced. These by-products could potentially cause unpredictable side effects.
● Drug Absorption: If drug metabolism in the intestine occurs rapidly, it may affect drug absorption, making it difficult to achieve effective concentrations of the drug.
● Individual Variability: Due to differences in the types and quantities of intestinal bacteria in individual patients, there can be variations in drug response even when the same drug is administered.
● BCS classification of drug (Class I to IV) : Over the last couple of years, there has been a tendency for newly developed drugs to be characterized by low solubility and/or permeability, thereby belonging to BCS class II, or III or IV drugs, but hardly to BCS class I drugs, which are highly soluble and highly permeable drugs (Ku, 2008).
This tendency is mainly caused by the increased structural complexity of newly designed drugs. As a consequence, the solubility and absorption of these drugs is impaired, resulting in prolonged residence in the gastrointestinal tract after oral administration and subsequently resulting in higher drug concentrations in the colon. However, not only can orally administered drugs end up in the colon, but also intravenously administered drugs can reach the intestines and colon via biliary excretion, such as after hepatic glucuronidation.
These factors have the potential to influence the selection of appropriate treatment for individual patients and the evaluation of the efficacy and safety of drugs. Therefore, the development of personalized pharmaceutical treatments for individual patients and drug design considering the impact of intestinal bacteria may shape the future direction of medicine.
TNO has a wide range of available tools to increase understanding of the impact and interaction of the microbiome on human health. This is key to determining how various diseases change the body, and how various drugs can impact that change.
Animation of targeted identification of compounds to restore a balanced microbiome.
Each i-screen can simultaneously be exposed to a variety of different conditions, thereby allowing multiple compounds or concentrations to be tested at once. In all cases, potential metabolites can be identified in the earliest stages of development, with human accuracy, without the need for regulatory processing. TNO can also put its microbiome expertise to work to conduct extensive toxicology testing and analysis. Using a variety of techniques and platforms, e.g.Ex vivo InTESTine model we can help investigate the interplay between microbiota and host, and the extent to which certain microbiota have a toxic effect on the human body.
I-Screen; Technology and Application
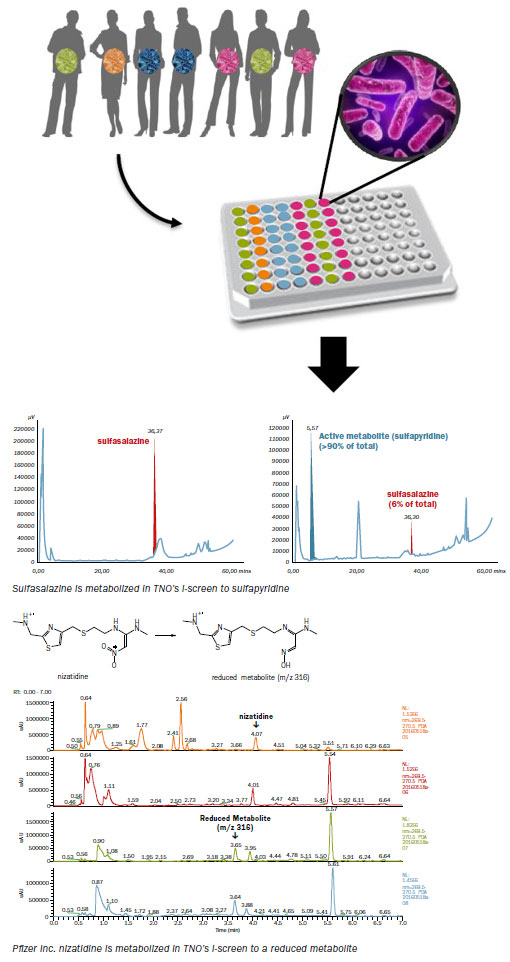 Human gut microbiota are cultured anaerobically in a multi-well system to mimic the human intestinal in vivo metabolism. This allows for determining the potential of intestinal microbiota to metabolize drug candidates. The starting population of microbiota in the I-screen can be a standardized ex-vivo intestinal microbiota pool collected from either healthy volunteers, or for example obese, or lean adults or even children. This would confer the inherent differences found in each individual’s specific gut microflora.
I-screen enables rapid identification of drugs that are susceptible to metabolism by intestinal microbiota. Additionally, it can be used to identify unknown metabolites transformed by intestinal microbiota. Normally, human metabolite profiling and identification data are generated during Phase II and/ or III of clinical trials. Human metabolites can thus be identified in preclinical stages, thereby de-risking and accelerating the
drug development process. Each I-screen can be simultaneously exposed to a large number of different conditions, allowing for cost effective screening of interesting candidate drugs.
Human gut microbiota are cultured anaerobically in a multi-well system to mimic the human intestinal in vivo metabolism. This allows for determining the potential of intestinal microbiota to metabolize drug candidates. The starting population of microbiota in the I-screen can be a standardized ex-vivo intestinal microbiota pool collected from either healthy volunteers, or for example obese, or lean adults or even children. This would confer the inherent differences found in each individual’s specific gut microflora.
I-screen enables rapid identification of drugs that are susceptible to metabolism by intestinal microbiota. Additionally, it can be used to identify unknown metabolites transformed by intestinal microbiota. Normally, human metabolite profiling and identification data are generated during Phase II and/ or III of clinical trials. Human metabolites can thus be identified in preclinical stages, thereby de-risking and accelerating the
drug development process. Each I-screen can be simultaneously exposed to a large number of different conditions, allowing for cost effective screening of interesting candidate drugs.
Example 1: Intestinal microbiota driven metabolism of Sulfasalazine
TNO has successfully applied the I-screen to demonstrate that Sulfasalazine in the I-screen system is metabolized as has been shown in vivo. Sulfasalazine is designed specifically to be metabolized by human intestinal microbiota to the 5-aminosalicylic acid that has therapeutic effect. However, clinical studies demonstrated that sulfasalazine is mainly metabolized to Sulfapyridine by the intestinal bacteria, which give rise to side-effects. In this example, microbiota from healthy adults were exposed to 100 μM sulfasalazine. Samples were taken after 0 and 24 hrs. and were analyzed by HPLC.
Example 2: Intestinal microbiota driven metabolism of Nizatidine, in collaboration with Pfizer Inc.
In a second example, TNO has successfully applied the i-screen in the investigation of Nizatidine, a pharmaceutical compound that is known to be metabolized by human intestinal microbiota. After the incubation period, the analysis of the samples was performed by Pfizer Inc. using high resolution mass spectrometry. The microbiota from healthy adults were exposed to 50 μM nizatidine and samples taken after 0, 6, 24 and 48 hrs, clearly demonstrated the metabolic capacity of gut microbiota.
Technolgies for Analysis
● Microbiome composition
> 16S or metagenome
> ITS Fungal composition
> Anti-biotic resistance
> Viability
● Microbiome Function
> Transcriptome
> Metabolome
> SCFA analysis
> Microbiome metabolites
> Drug metabolites
> Microscopy (fluorescence)
● Physiology of Host
> Blood biomarkers
> Metabolites
> Host-microbiome
> Interactions
I-screen Webinar information
On Tuesday, November 9, 2021 TNO webinar In vitro microbiome and ex vivo intestinal models entitled "WHAT'S NEW IN IN VITRO MICROBIOME AND EX VIVO INTESTINAL MODELS FOR GUT HEALTH" took place.
Program: You can watch this webinar on demand via this link:
Publication
Drug Metabolism and Disposition, March 2024
Gut Microbiome Integration in Drug Discovery and Development of Small Molecules
Patrick Jimonet, Celine Druart, Stephanie Blanquet-Diot, Lilia Boucinha, Stephanie Kourula, Francoise Le Vacon, Sylvie Maubant, Sylvie Rabot, Tom Van de Wiele, Frank Schuren, Vincent Thomas, Bernard Walther, Michael Zimmermann; Medicen Microbiome Drug Metabolism Working Group
Microbiome Research Report 2024
A host-microbial metabolite interaction gut-on-a-chip model of the adult human intestine demonstrates beneficial effects upon inulin treatment of gut microbiome
Joanne M. Donkers, Maria Wiese, Tim J. van den Broek, Esmee Wierenga, Valeria Agamennone, Frank Schuren and Evita van de Steeg.
Drug Metabolism and Disposition August 29, 2018,
An ex vivo fermentation screening platform to study drug metabolism by human gut microbiota
Evita van de Steeg, Frank H.J. Schuren, R. Scott Obach, Claire van Woudenbergh, Gregory S. Walker, Margreet Heerikhuisen, Irene H.G. Nooijen and Wouter H.J. Vaes
BMC Med Genomics. June 2014
A systems biology approach to understand the pathophysiological mechanisms of cardiac pathological hypertrophy associated with rosiglitazone.
Verschuren L, Radonjic M, Wielinga PY, Kelder T, Kooistra T, van Ommen B, Kleemann R.
Best Pract Res Clin Gastroenterol. Feb 2013.
Ex vivo systems to study host-microbiota interactions in the gastrointestinal tract.
Roeselers G, Ponomarenko M, Lukovac S, Wortelboer HM
PLoS One. Feb 2013
Differential effects of drug interventions and dietary lifestyle in developing type 2 diabetes and complications: a systems biology analysis in LDLr-/- mice.
Radonjic M, Wielinga PY, Wopereis S, Kelder T, Goelela VS, Verschuren L, Toet K, van Duyvenvoorde W, van der Werff van der Vat B, Stroeve JH, Cnubben N, Kooistra T, van Ommen B, Kleemann R.
Pharmacol Res. Dec 2012.
The human gastrointestinal microbiota--an unexplored frontier for pharmaceutical
Roeselers G, Bouwman J, Venema K,Montijn R.
Pharmacogenet Genomics. Dec 2012.
Systems biology analysis unravels the complementary action of combined rosuvastatin and ezetimibe therapy
Verschuren L, Radonjic M, Wielinga PY, Kelder T, Kooistra T, van Ommen B, Kleemann R.
Download; Posters, Technical Info
● TNO I-Screen (Brochure)
●I-Screen: An Ex Vivo Human Microbiome Platform to study Microbiome Induced Reverse Metabolism of Metabolites Back to Parent.(Poster at ISSX-2019)
● In vitro platform to study human gut microbiota induced drug metabolism and molecular transformations (Poster)
●The role of fungi in irritable bowel syndrome (IBS): new insights in human disease development(Poster)
●On Tuesday, November 9, 2021 TNO's webinar In vitro microbiome and ex vivo intestinal models entitled "WHAT'S NEW IN IN VITRO MICROBIOME AND EX VIVO INTESTINAL MODELS FOR GUT HEALTH" took place.
You can watch this webinar on demand via this link:

